-Search query
-Search result
Showing 1 - 50 of 69 items for (author: huang & rh)

EMDB-42820: 
SARS-CoV-2 5' proximal stem-loop 5 and 6 with SL5b extended
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R
EMDB-42816: 
SARS-CoV-1 5' proximal stem-loop 5
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

PDB-8uyp: 
SARS-CoV-1 5' proximal stem-loop 5
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

EMDB-42801: 
BtCoV-HKU5 5' proximal stem-loop 5, conformation 1
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

EMDB-42802: 
BtCoV-HKU5 5' proximal stem-loop 5, conformation 3
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

EMDB-42805: 
BtCoV-HKU5 5' proximal stem-loop 5, conformation 2
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R
EMDB-42808: 
BtCoV-HKU5 5' proximal stem-loop 5, conformation 4
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R
EMDB-42809: 
MERS 5' proximal stem-loop 5, conformation 1
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

EMDB-42810: 
MERS 5' proximal stem-loop 5, conformation 2
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

EMDB-42811: 
MERS 5' proximal stem-loop 5, conformation 3
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R
EMDB-42818: 
SARS-CoV-2 5' proximal stem-loop 5
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

EMDB-42819: 
SARS-CoV-2 5' proximal stem-loop 5 and 6 with SL5c extended
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R
EMDB-42821: 
SARS-CoV-2 5' proximal stem-loop 5 and 6
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R
PDB-8uye: 
BtCoV-HKU5 5' proximal stem-loop 5, conformation 1
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

PDB-8uyg: 
BtCoV-HKU5 5' proximal stem-loop 5, conformation 2
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

PDB-8uyj: 
BtCoV-HKU5 5' proximal stem-loop 5, conformation 4
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

PDB-8uyk: 
MERS 5' proximal stem-loop 5, conformation 1
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

PDB-8uyl: 
MERS 5' proximal stem-loop 5, conformation 2
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

PDB-8uym: 
MERS 5' proximal stem-loop 5, conformation 3
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

PDB-8uys: 
SARS-CoV-2 5' proximal stem-loop 5
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

EMDB-42803: 
HCoV-229E 5' proximal stem-loop 5
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R
EMDB-42813: 
HCoV-NL63 5' proximal stem-loop 5, conformation 1
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

EMDB-42814: 
HCoV-NL63 5' proximal stem-loop 5, conformation 2
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R
EMDB-29301: 
Neurotensin receptor allosterism revealed in complex with a biased allosteric modulator
Method: single particle / : Krumm BE, Diberto JF, Olsen RHJ, Kang H, Slocum ST, Zhang S, Strachan RT, Fay JF, Roth BL

EMDB-29302: 
CryoEM structure of Go-coupled NTSR1 with a biased allosteric modulator
Method: single particle / : Krumm BE, DiBerto JF, Olsen RHJ, Kang H, Slocum ST, Zhang S, Strachan RT, Fay JF, Roth BL

EMDB-29303: 
CryoEM structure of Go-coupled NTSR1
Method: single particle / : Krumm BE, DiBerto JF, Olsen RHJ, Kang H, Slocum ST, Zhang S, Strachan RT, Fay JF, Roth BL

EMDB-26767: 
The 2.19-angstrom CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - Complex minus stalk
Method: single particle / : Grinter R, Venugopal H, Kropp A, Greening C

EMDB-26801: 
The 1.67 Angstrom CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - catalytic dimer (Huc2S2L)
Method: single particle / : Grinter R, Venugopal H, Kropp A, Greening C

EMDB-26802: 
The CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - Full complex focused refinement of stalk
Method: single particle / : Grinter R, Venugopal H, Kropp A, Greening C
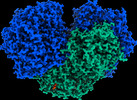
EMDB-27661: 
The 1.52 angstrom CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - catalytic dimer (Huc2S2L)
Method: single particle / : Grinter R, Venugopal H, Kropp A, Greening C

PDB-7utd: 
The 2.19-angstrom CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - Complex minus stalk
Method: single particle / : Grinter R, Venugopal H, Kropp A, Greening C

PDB-7uur: 
The 1.67 Angstrom CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - catalytic dimer (Huc2S2L)
Method: single particle / : Grinter R, Venugopal H, Kropp A, Greening C

PDB-7uus: 
The CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - Full complex focused refinement of stalk
Method: single particle / : Grinter R, Venugopal H, Kropp A, Greening C

PDB-8dqv: 
The 1.52 angstrom CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - catalytic dimer (Huc2S2L)
Method: single particle / : Grinter R, Venugopal H, Kropp A, Greening C

EMDB-27966: 
CryoEM structure of miniGq-coupled hM3Dq in complex with DCZ
Method: single particle / : Zhang S, Fay JF, Roth BL

EMDB-27967: 
CryoEM structure of miniGo-coupled hM4Di in complex with DCZ
Method: single particle / : Zhang S, Fay JF, Roth BL
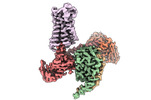
EMDB-27968: 
CryoEM structure of miniGq-coupled hM3Dq in complex with CNO
Method: single particle / : Zhang S, Fay JF, Roth BL

EMDB-27969: 
CryoEM structure of miniGq-coupled hM3R in complex with Iperoxo
Method: single particle / : Zhang S, Fay JF, Roth BL
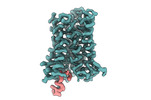
EMDB-27970: 
CryoEM structure of miniGq-coupled hM3R in complex with iperoxo (local refinement)
Method: single particle / : Zhang S, Fay JF, Roth BL

EMDB-27779: 
Structure of the SARS-CoV-2 spike glycoprotein S2 subunit
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

PDB-8dya: 
Structure of the SARS-CoV-2 spike glycoprotein S2 subunit
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-27752: 
CryoEM structure of Gq-coupled MRGPRX1 with peptide agonist BAM8-22
Method: single particle / : Liu Y, Cao C, Fay JF, Roth BL

EMDB-27753: 
CryoEM structure of Gq-coupled MRGPRX1 with peptide ligand BAM8-22 and positive allosteric modulator ML382
Method: single particle / : Liu Y, Cao C, Fay JF, Roth BL
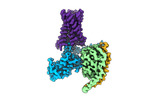
EMDB-27754: 
CryoEM structure of Gq-coupled MRGPRX1 with ligand Compound-16
Method: single particle / : Liu Y, Cao C, Fay JF, Roth BL

EMDB-24944: 
Damaged 70S ribosome with PrfH bound
Method: single particle / : Tian Y, Zeng F, Raybarman A, Carruthers A, Li Q, Fatma S, Huang RH

EMDB-24945: 
Intact 70S ribosome without PrfH bound
Method: single particle / : Tian Y, Zeng F, Raybarman A, Carruthers A, Li Q, Fatma S, Huang RH

EMDB-30947: 
Cryo EM structure of a Na+-bound Na+,K+-ATPase in the E1 state
Method: single particle / : Guo YY, Zhang YY, Yan RH, Huang BD, Ye FF, Wu LS, Chi XM, Zhou Q

EMDB-30948: 
Cryo EM structure of a K+-bound Na+,K+-ATPase in the E2 state
Method: single particle / : Guo YY, Zhang YY, Yan RH, Huang BD, Ye FF, Wu LS, Chi XM, Zhou Q

EMDB-30949: 
Cryo EM structure of a Na+-bound Na+,K+-ATPase in the E1 state with ATP-gamma-S
Method: single particle / : Guo YY, Zhang YY, Yan RH, Huang BD, Ye FF, Wu LS, Chi XM, Zhou Q

EMDB-31249: 
S protein of SARS-CoV-2 in complex with GW01
Method: single particle / : Shen YP, Zhang YY, Yan RH, Li YN, Zhou Q
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
